Guangzhou Dongsheng Biotech Co., Ltd |
|
Verified Suppliers
|
|
【Product Name】
Fast DNA Library Plus Prep Kit for MGI
【Cat. No./Spec.】
KM004-A/24 rxns; KM004-B/96 rxns; sample sack/ 6 rxns
【Product Description】
Aiming at MGI high-throughput sequencing platform, this kit provides a convenient and universal DNA library construction scheme in one tube. It combines fragmentation, end repair and A-Tailing into one step, greatly shortening the time of library construction and reducing the error caused by tedious steps. After fragmentation and end preparation, the product can be directly ligated with adapter without additional purification, and the subsequent procedure is the same as that of #KM001 Fast DNA Library Prep Kit for MGI. Complete library quantification can be performed by dsDNA fluorescent dye method (e.g., Thermo Qubit Flex Fluorometer) or absolute quantification PCR after diluting the library to an appropriate concentration.
【Sample Type】
Table 1 Recommended Inputs for Common DNA
| Application | Sample Type | Recommended Amount |
| Whole genome sequencing | High quality complex genomes | 50ng-1μg |
| Target capture sequencing of whole exome | High quality complex genomes | 10ng-1μg |
| Target capture sequencing of whole genome | FFPE DNA | ≥50ng |
| Whole genome sequencing | Microbial genome | 1ng-1μg |
【Storage Condition & Shelf Life】
All reagents should be stored at -20°C. Ligation Buffer is normal for crystals to precipitate at low temperatures, it should be balanced to room temperature before use. The product is valid for 12 months.
【Components】
| Component | 24 rxns | 96 rxns |
| FEP Buffer | 120 μl | 480 μl |
| FEP Enzyme Mix | 240 μl | 2×480 μl |
| Fast DNA Ligase | 120 μl | 2×240 μl |
| Fast Ligation Buffer | 600 μl | 4×600 μl |
| 2× HIFI Library PCR Master Mix | 600 μl | 4×600 μl |
| Primer Mix for MGI * | 120 μl | 480 μl |
| Neutralization Buffer | 120 μl | 480 μl |
*FEP Buffer is the fragmentation and end preparation reaction buffer. FEP Enzyme Mix is a mixture of enzyme related to fragmentation and end preparation.
* If there are more than one sample, #KM002 and #KM003 adapter primer mix is recommended. This kit provides a set of primers, the primer sequence is as follows:
5’-TGTGAGCCAAGGAGTTG-3’
5’-GAACGACATGGCTACGA-3’
Note:recommended selection beads: #NC1011 GDSPure DNA Selection Magbeads or AMPure XP beads.
【Notes】
1. We offer two types of Universal Adapter primers set (GDS Adapter, #KM002 and #KM003, purchased separately), but customers can also choose from other manufacturers or synthesize their own Adapter for the MGI sequencing platform. Too much Adapter will lead to the formation of Adapter dimer, and insufficient Adapter will lead to low library output. Therefore, appropriate Adapter concentration determines the concentration and quality of library. The recommended adapter concentrations for different amounts of DNA input are shown in the following table:
Table 2 Recommended Use Concentrations of Adapter
| DNA Input | Recommended Conc. for Adapter | Adapter:Insert Mole Ratio | GDS Adapter Dilution Degrees* |
| 1μg | 10μM | 10:1 | No dilution |
| 500ng | 10μM | 20:1 | No dilution |
| 250ng | 10μM | 40:1 | No dilution |
| 100ng | 7.5μM | 100:1 | 3:4 |
| 50ng | 5μM | 200:1 | 1:2 |
| 25ng | 2.5μM | 200:1 | 1:4 |
| 1ng | 1μM | 200:1 | 1:10 |
* Expressed as the volume ratio of adapter to diluent
2. The enzyme used in 2× HIFI Library PCR Master Mix is a B family DNA polymerase, which has 5 '-3' polymerase and 3 '-5' exonuclease activities, but lacks 5 '-3' exonuclease activities. It has high fidelity and homogeneity, and strong sustainable synthesis ability. Strict control of the number of amplification cycles is particularly important for library output. The following table shows the recommended number of amplification cycles corresponding to different amounts of DNA input:
Table 3 Recommended Number of Amplification Cycles Corresponding to Different Sample Inputs
| Input DNA | Recommended Number of Amplification Cycles | |
| 100ng Library | 1μg Library | |
| 1μg | 0 | 2-5 |
| 500ng | 0 | 2-5 |
| 250ng | 1-3 | 5-7 |
| 100ng | 2-4 | 6-8 |
| 50ng | 4-6 | 8-10 |
| 25ng | 5-7 | 9-12 |
| 10ng | 7-9 | 11-13 |
| 5ng | 9-11 | 13-14 |
| 2.5ng | 10-12 | 14-16 |
| 1ng | 11-13 | 15-17 |
Note: 1. The above table shows the test results using 150bp standard DNA, which is for reference only.
2. If incomplete connectors are used, a minimum number of cycles (1-3) should be amplified to obtain a complete library.
3. If the quality of the input DNA is poor, or the size selection is carried out during the library construction, the number of amplification cycles should be appropriately increased.
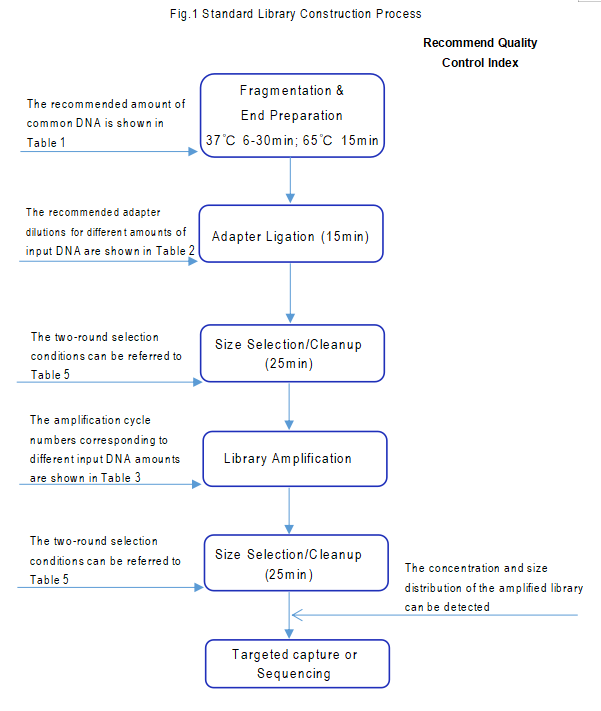
【Standard Library Construction Process】
Fragmentation and End Repair
1. Determine the solvent composition of template DNA, if no EDTA, proceed directly to Step 2; If EDTA is contained, 2.2× magnetic beads should be used for purification, or a corresponding volume of Neutralization Buffer should be added according to the content of EDTA in the following table for Neutralization:
| EDTA Conc. | Volume of Neutralization Buffer |
| 1mM | 5 μl |
| 0.8mM | 4 μl |
| 0.6mM | 3 μl |
| 0.5mM | 2.5 μl |
| 0.4mM | 2 μl |
| 0.2mM | 1 μl |
| 0.1mM | 0.5 μl |
| <0.1mM | 0 μl |
2. Prepare the following reaction in a 200 μl PCR tube:
| Reagents | Volume |
| Input DNA | X μl |
| FEP Buffer | 5 μl |
| Neutralization Buffer | Y μl |
| ddH2O | To 65 μl |
3. Add 10 μl FEP Enzyme Mix to the above system, blow evenly, centrifuge briefly, and immediately put into PCR instrument for the following reaction:
| Temperature | Time |
| 20°C | 15min |
| 37°C | Refer to Table 4 |
| 65°C | 15min |
| 4°C | ∞ |
Table 4 Holding Time Required to Obtain Libraries of Different Sizes
| Fragment Size | Time |
| 150bp | 20-30min |
| 250bp | 15-20min |
| 350bp | 10-15min |
| 550bp | 6-10min |
Adapter Ligation
1. Proceed with the ligation reaction as soon as possible after fragmentation and end preparation.
2. Dilute the adapter according to Table 2.
3. Prepare the following reaction system:
| Reagents | Volume |
| above products | 50 μl |
| Fast Ligation Buffer | 25 μl |
| Fast DNA Ligase | 5 μl |
| Adapter X for MGI | 5 μl |
| ddH2O | 15 μl |
| Total | 100 μl |
4. Vortex gently and spin down briefly to mix well, centrifuge briefly and collect all the liquid to the bottom of the tube.
5. Perform the following reaction in a thermal cycler:
| Temperature | Time |
| 20°C | 15min |
| 4°C | ∞ |
Recommended Solution for PCR Cleanup/Size Selection (the specific magnetic bead volume should be adjusted according to the actual sample size)
1. Prepare 100 μl ligation products into an appropriate centrifuge tube.
2. Add 100 μl of resuspended DNA selection magnetic beads to the sample. Gently blow with a pipette for 10 times (or vortex for 30 s). Incubate samples for 5 min at room temperature.
3. Place the tube on an appropriate magnetic rack to separate the beads from the supernatant. When the solution is clear, carefully remove and discard the supernatant with a pipette (do not discard beads).
4. Add 200 μl of 80% freshly prepared ethanol to the tube while in the magnetic rack. Incubate at room temperature for 30 s, and then carefully remove and discard the supernatant (do not disturb beads).
5. Repeat Step 4 once for a total of two washes.
Note: Be sure to remove all visible liquid after the second wash.
6. Air dry the beads until the surface of the magnetic beads has no obvious gloss while the tube is on the magnetic rack with the lid open.
Note: Do not overdry the beads, this may result in lower recovery of DNA. When the beads start to crack, they are too dry.
7. Remove the tube from the magnetic rack. Add 22 μl elution buffer (10mM Tris-HCl, pH8.0-8.5) to the tube. Mix well by pipetting up and down at least 10 times or on a vortex mixer for 30 s. Incubate for 3-5 min at room temperature.
8. Place the tube on the magnetic rack. After 5 min (or when the solution is clear), transfer 20 μl supernatant to a new tube. The selection is completed, and the selected DNA can be used for subsequent experiments or stored at -20°C for a long time.
Library Amplification
1. Prepare the following reaction in a PCR tube:
| Reagents | Volume |
| Ligation products after cleanup or size selection | 20 μl |
| 2× HIFI Library PCR Master Mix | 25 μl |
| Primer mix for MGI | 5 μl |
| Total | 50 μl |
2. Vortex gently and spin down briefly to mix well, centrifuge briefly and collect all the liquid to the bottom of the tube.
3. Perform the following reaction in a thermal cycler:
| Temperature | Time | Cycle Number |
| 95°C | 3min | 1 |
| 98°C | 20sec |
Select appropriate number of cycles according to Table 3 |
| 60°C | 15sec | |
| 72°C | 30sec | |
| 72°C | 5min | 1 |
| 4°C | ∞ | - |
Recommended Solution for PCR Cleanup/Size Selection (the specific magnetic bead volume should be adjusted according to the actual sample size)
1. Prepare 50 μl ligation products into an appropriate centrifuge tube.
2. Add 45 μl of resuspended DNA selection magnetic beads to the sample. Gently blow with a pipette for 10 times (or vortex for 30 s). Incubate samples for 5 min at room temperature.
3. Place the tube on an appropriate magnetic rack to separate the beads from the supernatant. When the solution is clear, carefully remove and discard the supernatant with a pipette (do not discard beads).
4. Add 200 μl of 80% freshly prepared ethanol to the tube while in the magnetic rack. Incubate at room temperature for 30 s, and then carefully remove and discard the supernatant (do not disturb beads).
5. Repeat Step 4 once for a total of two washes.
Note: Be sure to remove all visible liquid after the second wash.
6. Air dry the beads until the surface of the magnetic beads has no obvious gloss while the tube is on the magnetic rack with the lid open.
Note: Do not overdry the beads, this may result in lower recovery of DNA. When the beads start to crack, they are too dry.
7. Remove the tube from the magnetic rack. Add 22 μl elution buffer (10mM Tris-HCl, pH8.0-8.5) to the tube. Mix well by pipetting up and down at least 10 times or on a vortex mixer for 30 s. Incubate for 3-5 min at room temperature.
8. Place the tube on the magnetic rack. After 5 min (or when the solution is clear), transfer 20 μl supernatant to a new tube. The selection is completed, and the selected DNA can be stored at 2-8°C for 1-2 weeks or stored at -20°C for a long time.
【Appendix】Recommended Scheme for Double-Sided Selection
If double-round selection is required, we provide the following scheme to select the appropriate magnetic bead volume according to the expected library size. The size selection can be performed before end repair or after amplification. Two or more double-round selection will greatly reduce the library yield.
Fill the library volume in the table below to 100 μl. Select the volume of magnetic beads in two rounds according to the expected library size. And carry out the selection operation according to the following instructions.
Table 5 Recommended Amount of Magnetic Beads for Double-Round Selection
| Expected Library Size | 150bp | 200bp | 250bp | 300bp | 400bp | 500bp | 600bp | 700bp | |
| Volume of Beads(μl) | Round 1 | 100 | 90 | 80 | 70 | 60 | 55 | 50 | 45 |
| Round 2 | 30 | 20 | 20 | 20 | 20 | 15 | 15 | 15 | |
1. Fill the library volume to 100 μl in a 200μl PCR tube and labeled as A. Add a certain volume of Magnetic beads according to the Table 5 (Round 1) to the tube A. Gently blow with a pipette for 30 s. Incubate samples for 5 min at room temperature.
2. Place the tube A on an appropriate magnetic rack to separate the beads from the supernatant. When the solution is clear, carefully remove the supernatant to a new tube and label it as B. Discard beads.
3. Add a certain volume of Magnetic beads according to the Table 5 (Round 2) to the tube B. Gently blow with a pipette for 30 s. Incubate samples for 5 min at room temperature. Place the tube B on magnetic rack. When the solution is clear, carefully remove and discard the supernatant.
4. Add 200 μl of 80% freshly prepared ethanol to the tube B while in the magnetic rack. Incubate at room temperature for 30 s, and then carefully remove and discard the supernatant (do not disturb beads).
5. Repeat Step 6 once for a total of two washes.
Note: Be sure to remove all visible liquid after the second wash.
6. Air dry the beads until the surface of the magnetic beads has no obvious gloss while the tube B is on the magnetic rack with the lid open.
Note: Do not overdry the beads, this may result in lower recovery of DNA. When the beads start to crack, they are too dry.
7. Remove the tube B from the magnetic rack. Add 22 μl elution buffer to the tube. Mix well by pipetting up and down at least 10 times or on a vortex mixer for 30 s. Incubate for 3-5 min at room temperature.
Note: If targeted capture will not be performed, add elution buffer (10mM Tris-HCl, ph 8.0-8.5) for elution. Otherwise, sterilized ultrapure water should be used for elution.
For Research Use Only
GDSBio Is a high-tech enterprise focusing on research and development, production and sales of high-quality life science products. The company has a complete product line, with PCR technology as the core, focusing on general PCR, fluorescence quantitative PCR, NGS storage, nucleic acid electrophoresis and other molecular biology technologies, and has developed molecular research reagents, molecular in vitro diagnostic raw materials, nucleic acid extraction and detection reagents and other products.

